E-mail: hbruce (at) mbl.edu
Research Interests
I use arthropod appendages to ask questions about the origin and evolution of novel structures, and how genetic networks evolve over vast phylogenetic distances on the order of hundreds of millions of years.
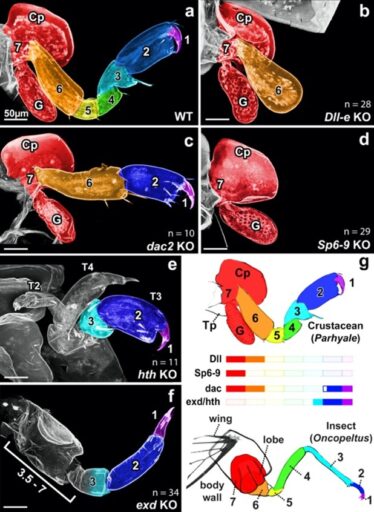
Fig. 1. CRISPR-Cas9 knockout of leg patterning genes in the crustacean Parhyale reveal a one-to-one correspondence between insect and crustacean distal leg segments.
I am interested in how genetic networks produce morphological diversity. This relates to the origin of novel structures, which are those that don’t seem to be derived from (homologous to) any structure in the ancestor (Müller and Wagner, 1991). Co-option of genetic pathways by unrelated tissues is often invoked to explain the origin of novel structures. However, my research into arthropod novel structures suggests an exciting alternative: novel structures may evolve from existing structures (serial homologs) that persist cryptically in other lineages. Many unexpected structures may thus be related and far more ancient and evolvable than currently believed. This has deep implications for how we assume genetic networks evolve, as it suggests that the wholesale swapping of networks, as assumed by co-option, may not be a common mechanism of network evolution, and we should instead explore models where networks are homologous but with millions of years of systems drift.
As a Ph.D. student in Nipam Patel’s lab at the University of California, Berkeley, I discovered a solution to one of the most contentious problems in evolution: the origin of insect wings. To do so, I performed CRISPR-Cas9 knockout of leg patterning genes in the crustacean Parhyale hawaiensis and compared my knockout phenotypes to those previously published in insects (Fig. 1). This comparison suggested that insect wings evolved from a multi-functional lobe (exite) on the ancestral crustacean leg. Thus, insect wings are not novel structures, as argued by many previous authors, but instead evolved from existing, ancestral structures (Bruce and Patel, 2020).
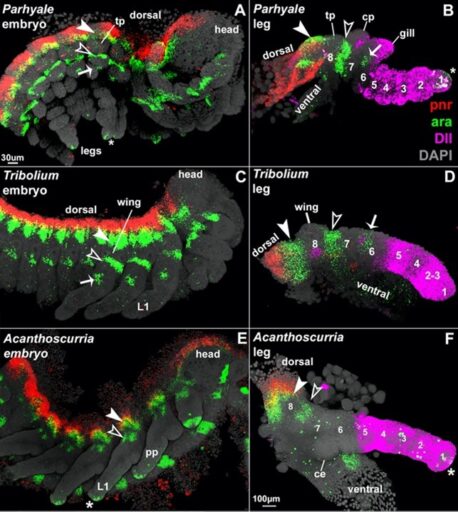
Fig. 2. In situ HCR of pnr (red) and ara (green) expression distinguish and homologize the proximal leg region while Dll (pink) patterns distal leg segments 1-5 in all arthropods.
As a Research Associate at the Marine Biological Laboratory, I extended this analysis to arachnids (Fig. 2), which revealed that all arthropod leg segments are patterned in a conserved manner with a simple one-to-one correspondence of segments (Bruce, 2022), much like the conserved Hox patterning system for the anterior-posterior axis. My straightforward coordinate system proved to be the key to answering over a century of speculation regarding the origins and relationships of the baroque horns, helmets, glands, and gills that decorate arthropods: surprisingly, all appear to be derived from ancestral exites. This breakthrough research has led to several national and international collaborations (Bruce, 2022).
To begin applying my proximal-distal coordinate system to the above enigmatic structures, I investigated another arthropod novel structure, the cape-like carapace of the crustacean, Daphnia (Shiga et al., 2017). By comparing the expression and function of genes from my model in Parhyale, Daphnia, and Tribolium (insect), I found that the carapace appears to be a greatly expanded exite, just like the insect wing. Incredibly, the exite that forms the carapace in Daphnia is still present in both Parhyale and Tribolium, but only as a small protrusion. Thus, rather than a novel structure resulting from gene co-option, the Daphnia carapace appears to have arisen from a shared, ancestral exite that persists in a cryptic state in other arthropod lineages (Bruce and Patel, 2022).
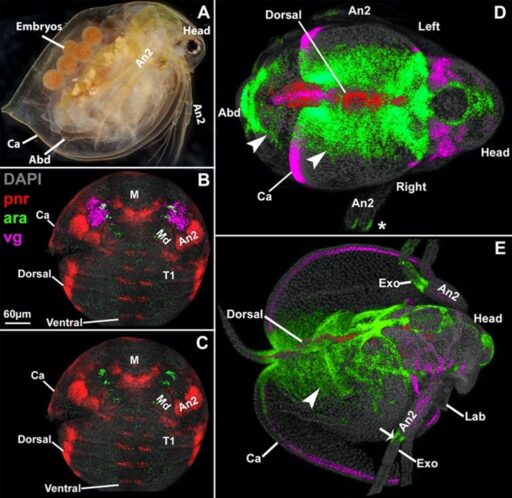
Fig. 3. The Daphnia carapace, like the insect wing and Parhyale tergal plate, expresses vestigial (vg, pink) around the edge, and extends from between two stripes of araucan (ara, green).
I next applied my coordinate system to insect abdominal outgrowths. This work suggested that all insects retain abdominal legs inherited from their crustacean ancestors, but only the proximal portion of the leg remains. These proximal leg nubs are visible in many insect embryos but degenerate to form the lateral body wall (pleurites and lateral tergum) of insects (Fig. 4). Importantly, these truncated abdominal legs appear to retain the ancestral ability to form multi-functional exites (gills, plates, etc). Thus, just as insect wings on the thorax evolved from an ancestral exite on a proximal leg segment that had been incorporated into the body wall (Bruce and Patel, 2020), so too did the various insect outgrowths on the abdomen, including gills, leaf-like structures used in camouflage, and colorful spines used for aposematic warning (Bruce and Patel, 2023).
Given that many seemingly novel structures in arthropods appear to be derived from ancestral structures that persist cryptically in intermediate lineages, this calls into question a main leg upon which the co-option literature rests. While my work is restricted to arthropods, cryptic persistence could be a phylogenetically widespread mechanism for the origin of novel structures. If so, it may be prudent for the evo-devo community to revise or redefine co-option (when defined as network swapping between non-homologous tissues).
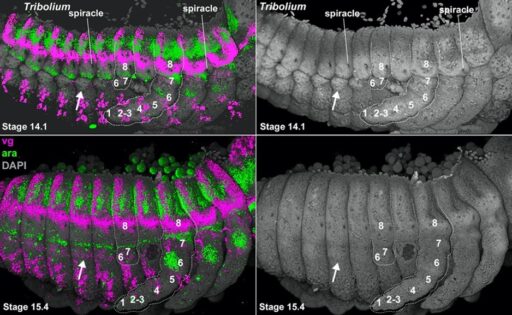
Fig. 4. The Daphnia carapace, like the insect wing and Parhyale tergal plate, expresses vestigial (vg, pink) around the edge, and extends from between two stripes of araucan (ara, green).
About Me
I was born and raised in the south Bay Area, near Starfleet headquarters, until I was 12 years old, when my parents moved to the backwater Class H desert planet of Cornville, Arizona. Although neither of my parents had attended Starfleet, I had always wanted to explore strange worlds, to seek out new life and new civilizations, and to boldly go where no one had gone before, so I decided to become a Starfleet cadet. I transferred to the tiny M-class planet of Yavapai College, and trained in the biological sciences under the tutelage of Captain Chris Breitmeyer and First Officer Jan Albright. For my birthday, they gave me the famous evodevo manual, Endless Forms Most Beautiful, by Sean Carroll. I was entranced by the idea that animals were constructed from networks of genetic circuits. In theory, the genetic architecture of development could be tweaked to create designer animals, such as reproductively-hindered, K-selected tribbles. Of course, the experimental foundations had to be established before this could become reality. Therefore, upon earning my Associate of Arts from Starfleet, I transferred to the M-class planet the University of Arizona, where I majored in Molecular and Cellular Biology.
My first mission was with Captain Lisa Nagy, of the USS Ilyanassa obsoleta. My mission was to determine whether the Hox gene post2, an orthologue of AbdB, played a role in the development and evolution of the mollusk shell, a morphological novelty. For my heroism at the University of Arizona, I was promoted to Lieutenant of Science, and transferred to my dream assignment on the major M-class planet, the University of California, Berkeley. There, I was Lieutenant of Hox Operations for two of the greatest Starfleet Officers of evo devo, Captain Nipam Patel of the mighty USS Parhyale hawaiensis, decorated for his glorious battle against countless developmentally important genes in non-model arthropods; and Captain Mike Eisen, a pioneer of the latest transcriptomics-class starship USS Drosophila melanogaster and fearless crusader of Open Access science publishing. My first mission was to characterize the Hox gene network and appendage diversity in the crustacean Parhyale hawiensis. Unfortunately, my mission would not have been completed in time before my shuttlecraft had to return, so I switched to a different mission. Fortunately, my second mission was a great success! I discovered a proximal-distal coordinate system for aligning all arthropod legs, which provided a framework for answering long-standing questions about the origins and relationships of arthropod outgrowths like horns, helmets, gills, and stink glands.
I am currently the First Officer of Captain Nipam Patel on the M-class planet Marine Biological Laboratory. I am also applying for evo-devo Captain positions, where I plan to command a fleet of four emerging model species-class starships, the USS Parhyale, the USS Tribolium, the USS Oxidus, and the USS Parasteatoda. With this fleet, I will explore the evolution of genetic networks over vast phylogenetic distances on the order of hundreds of millions of years.


